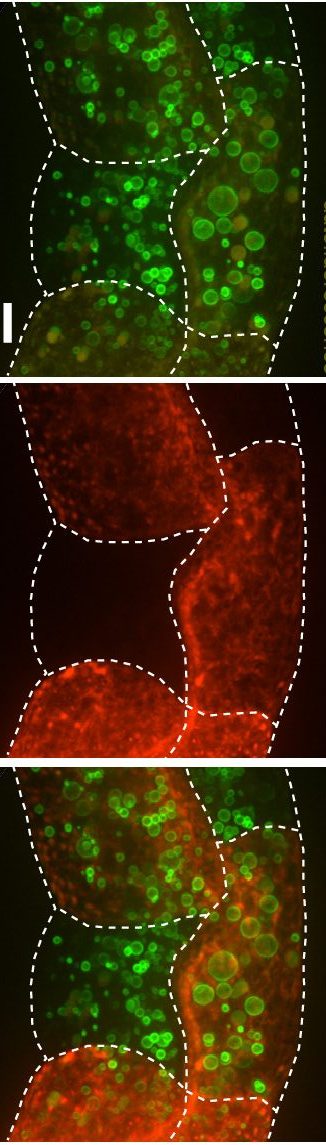
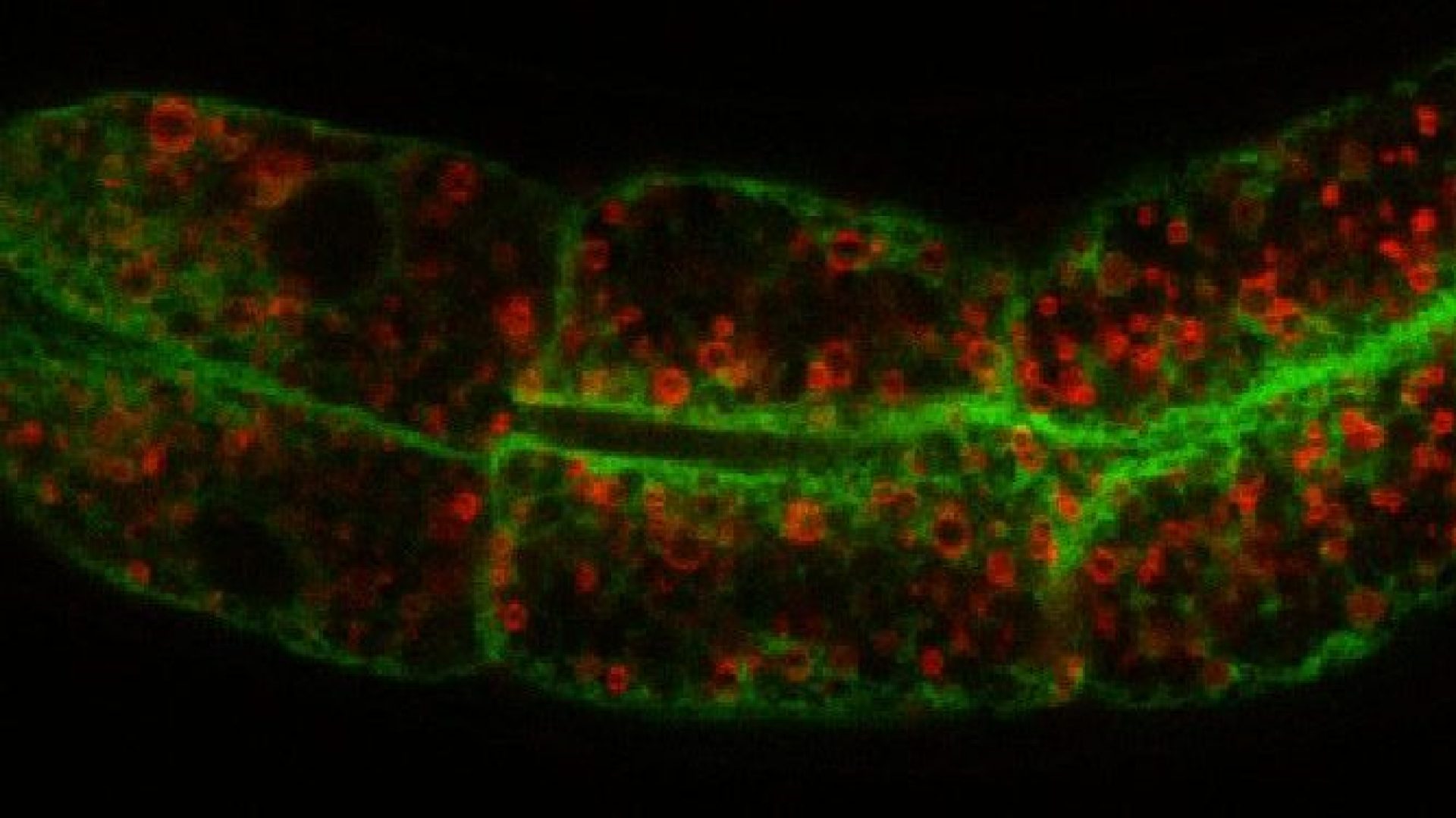
Regulation of cellular fat storage and mobilization
Our laboratory uses the nematode C. elegans as a discovery tool to elucidate the genetic pathways and molecular mechanisms that regulate food intake, fat storage and mobilization. We seek to address the following questions: (1) what are the regulatory mechanisms that govern lipid droplet size; (2) how do lipid droplets physically and functionally couple with other intracellular organelles; (3) what are the mechanisms for protein targeting and retention to lipid droplets? Most of our studies begin with large-scale forward genetic screens in C. elegans. We then employ genetics, lipid and protein biochemistry, proteomics, high-throughput sequencing, fluorescence and electron microscopy to address our questions at a molecular level in C. elegans and mammalian models.

Our team
Principle Investigator
- Ho Yi MAK
Technician
- Frances CHAN
Graduate Students
- Siwei HUANG (2019-)
- Lam Shing CHAN (2022-)
- Kaili DING (2024-)
- Songmin LEE (2024-)
- Chenwei SHI (2024-)
- Man Lung TSOI (2024-)
- Shing Hei HUNG (2025-)
- Ho Yin CHEUNG (2025-)
- Thomson HUSEIN (2025-)
Undergraduate students
- Seyeon KIM (2024-)
- Geonu KIM (2025-)
- Minseob SO (2025-)
Selected alumni
- Zhe CAO
- Postdoc @ Hotamisligil lab, Harvard Medical School
- Lidan ZENG
- Postdoc @ Wan lab, Emory University School of Medicine
- Yan LI
- Postdoc @ Scherer lab, UT Southwestern Medical Center
- Hok Chun MO
- Grad student @ Pagliarini lab, Washington University School of Medicine
Publications
Xie K, Liu Y, Li X, Zhang H, Zhang S, Mak HY, Liu P. Dietary S. maltophilia induces supersized lipid droplets by enhancing lipogenesis and ER-LD contacts in C. elegans. Gut Microbes. 2022; 14(1):2013762. doi: 10.1080/19490976.2021.2013762.
Zeng L, Li X, Preusch CB, He GJ, Xu N, Cheung TH, Qu J, Mak HY*. Nuclear receptors NHR-49 and NHR-79 promote peroxisome proliferation to compensate for aldehyde dehydrogenase deficiency in C. elegans. PLoS Genet. 2021; 17(7):e1009635. doi: 10.1371/journal.pgen.1009635.
Cao Z, Wang X, Huang X, Mak HY*. Are endoplasmic reticulum subdomains shaped by asymmetric distribution of phospholipids? Evidence from a C. elegans model system. Bioessays. 2021; 43(1):e2000199. doi: 10.1002/bies.202000199.
Cao Z, Mak HY*. Married at Birth: Regulation of Cellular Fat Metabolism by ER–Lipid Droplet Crosstalk. Contact. January 2020. doi:10.1177/2515256420934671
Na H, Zhang P, Chen Y, Zhu X, Liu Y, Liu Y, Xie K, Xu N, Yang F, Yu Y, Cichello S, Mak HY, Wang MC, Zhang H, Liu P. Identification of lipid droplet structure-like/resident proteins in Caenorhabditis elegans. Biochim Biophys Acta. 2015 Oct;1853(10 Pt A):2481-91.
Yang H, Vallandingham J, Shiu P, Li H, Hunter CP, Mak HY*. The DEAD Box Helicase RDE-12 Promotes Amplification of RNAi in Cytoplasmic Foci in C. elegans. Curr Biol. 2014; 24(8):832-8.
Krajniak J, Hao Y, Mak HY, Lu H. C.L.I.P.–continuous live imaging platform for direct observation of C. elegans physiological processes. Lab Chip. 2013 Aug 7;13(15):2963-71.
Klemm RW†, Norton JP†, Cole RA†, Li CS, Park SH, Crane MM, Li L, Jin D, Boye-Doe A, Liu TY, Shibata Y, Lu H, Rapoport TA, Farese RV Jr, Blackstone C, Guo Y*, Mak HY*. A conserved role for atlastin GTPases in regulating lipid droplet size. Cell Rep. 2013 May 30;3(5):1465-75.
Xu N, Zhang SO, Cole RA, McKinney SA, Guo F, Haas JT, Bobba S, Farese RV Jr, Mak HY*. The FATP1-DGAT2 complex facilitates lipid droplet expansion at the ER-lipid droplet interface. J Cell Biol. 2012 Aug 27;198(5):895-911.
Yang H, Zhang Y, Vallandingham J, Li H, Florens L, Mak HY*. The RDE-10/RDE-11 complex triggers RNAi-induced mRNA degradation by association with target mRNA in C. elegans. Genes Dev. 2012 Apr 15;26(8):846-56.
Hao Y, Xu N, Box AC, Schaefer L, Kannan K, Zhang Y, Florens L, Seidel C, Washburn MP, Wiegraebe W, Mak HY*. Nuclear cGMP-dependent kinase regulates gene expression via activity-dependent recruitment of a conserved histone deacetylase complex. PLoS Genet. 2011 May;7(5):e1002065.
Zhang SO, Trimble R, Guo F, Mak HY*. Lipid droplets as ubiquitous fat storage organelles in C. elegans. BMC Cell Biol. 2010 Dec 8;11:96.
Zhang SO, Box AC, Xu N, Le Men J, Yu J, Guo F, Trimble R, Mak HY*. Genetic and dietary regulation of lipid droplet expansion in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2010 Mar 9;107(10):4640-5.
Butcher RA, Ragains JR, Li W, Ruvkun G, Clardy J, Mak HY*. Biosynthesis of the Caenorhabditis elegans dauer pheromone. Proc Natl Acad Sci U S A. 2009 Feb 10;106(6):1875-9.